The human brain, a quest for simplification & understanding
The most complex object in the universe
The present approach to understanding and treating disorders of the brain is one of reverence to its complexity. Other than disorders with visible structural lesions, observing the organ has traditionally yielded little aid in explaining and treating neurological illnesses. Symptoms and behaviors are instead necessary proxies.
Indeed, the Human Connectome Project (HCP) confirmed what many suspected - the complexity of the brain really was beyond what we could humanly comprehend. Traditional views of the brain, such as the 52 Brodmann areas, simply failed to explain higher functions such as speech and cognition, and diseases such as mental illness.

Dive Deeper
Nullam id dolor id nibh ultricies vehicula ut id elit. Donec ullamcorper nulla non metus auctor.
Fusce dapibus, tellus ac cursusmmodo, tortor mauris condimentum nibh, ut fermentum massa justo sit amet risus. Cras justo odio, dapibus ac facilisis in, egestas eget quam. Nullam id dolor id nibh ultricies vehicula ut id elit.
The most complex object in the universe
The present approach to understanding and treating disorders of the brain is one of reverence to its complexity. Other than disorders with visible structural lesions, observing the organ has traditionally yielded little aid in explaining and treating neurological illnesses. Symptoms and behaviors are instead necessary proxies.
Indeed, the Human Connectome Project (HCP) confirmed what many suspected - the complexity of the brain really was beyond what we could humanly comprehend. Traditional views of the brain, such as the 52 Broddman areas, simply failed to explain higher functions such as speech and cognition, and diseases such as mental illness.
The brain as a data problem
With the HCP, nonetheless, came hope that we could begin to quantify and alphabetise the regions of the brain by function, not anatomy. By combining digital tractography and fMRI imaging, it is now possible to create individualised connectomic brain maps and draw forth millions more data points than we ever had available.
This, however, also generated more data than could practically be handled in many research settings and most clinical environments. There is now tension that using connectomic data to better understand and treat an individual’s brain is possible, but not practical.
Indeed, a growing number of ‘computational’ neuroscientists is a telling sign that the brain is better treated as a data problem in addition to a biological one, and for this approach most hospitals and neuroscience centers are ill-equipped.
Streamlining, standardizing, and supercharging your analytical pipeline
The aim of Omniscient’s software is to automate data analytics at scale, leaving clinicians and researchers with insights to better inform clinical decision making and research questions.
Our approach is one of:
Streamlining via dimension reduction - creating greater understanding by reducing the problem to its key variables
Standardizing via automating the labor intensive and error-prone steps
Supercharging via secure access to industrial-scale computational power in the cloud, allowing scans to be processed in an hour
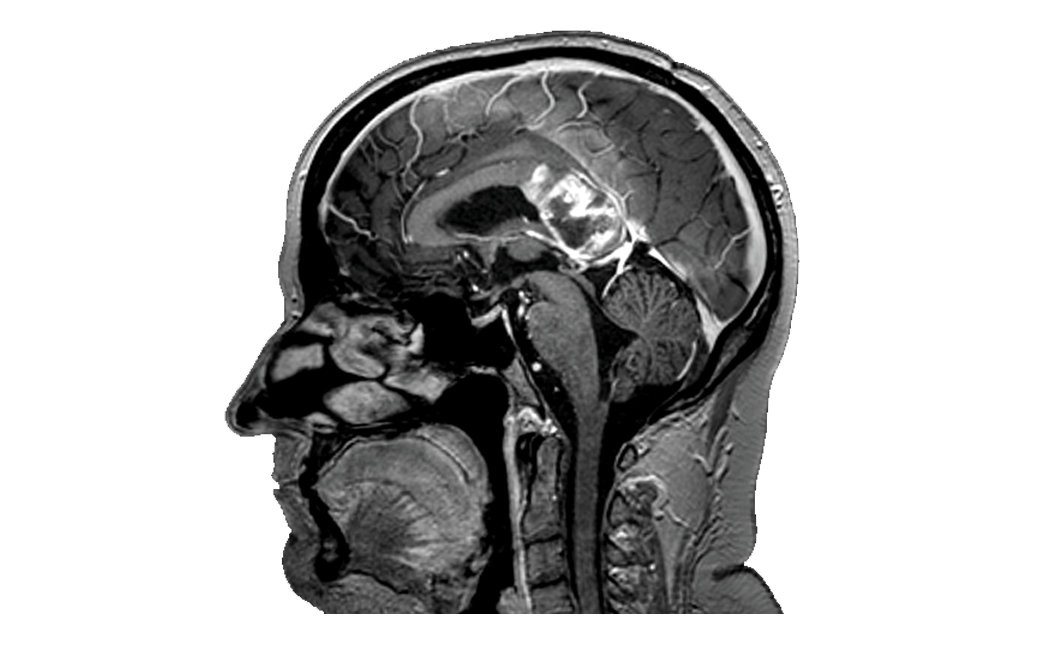
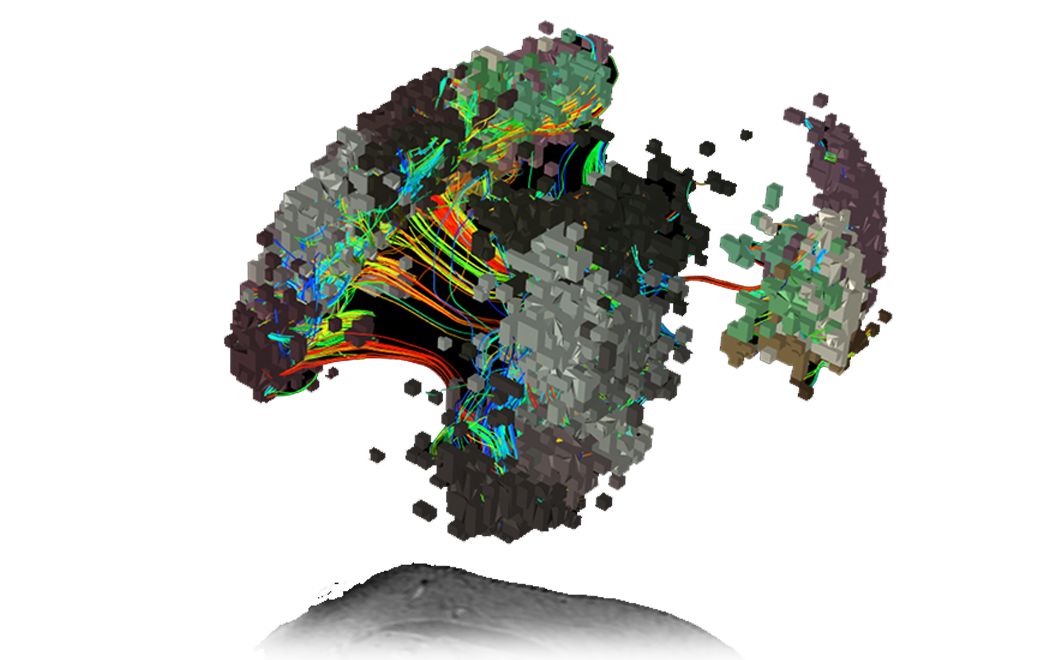
T1 Anatomical MRI images are visualised to co-register analysed data.
Traditionally, these images provided little insight into brain function. Note, this subject has a high grade butterfly glioma.
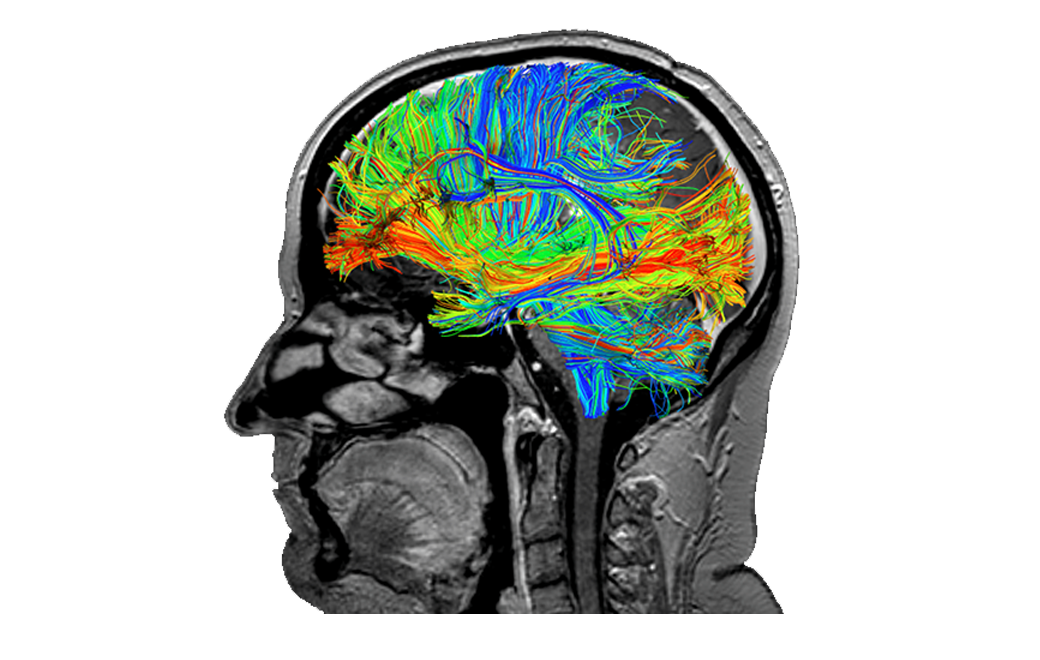
The structural connections of the brain are visualized through white matter tracts mapped with constrained spherical deconvolution (CSD) tractography.
Compared to standard diffusion tensor imaging (DTI), the use of CSD mitigates the impact of intra-voxel crossing of fibres.
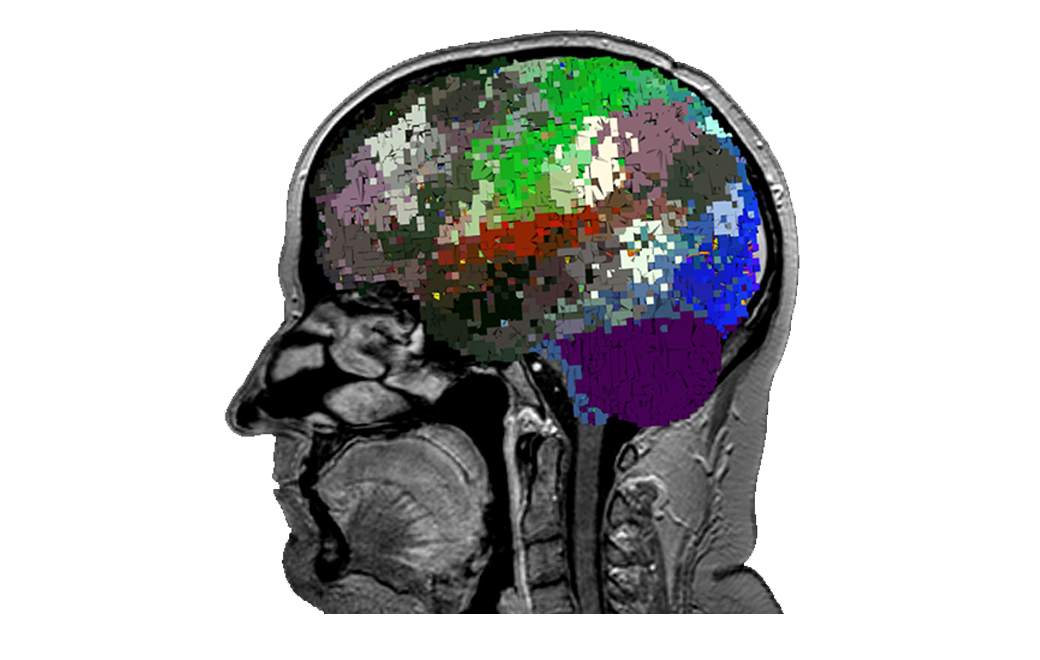
Using in-house technologies, the ‘parcellations’1 or functional areas of the cortex discovered and described through the Human Connectome Project are mapped out for this specific subject, even around the glioma.
1. Doyen, S., Nicholas, P., Poologaindran, A., Crawford, L., Young, I. M., Romero-Garcia, R., & Sughrue, M. E. (2021). Connectivity-based parcellation of normal and anatomically distorted human cerebral cortex. Human Brain Mapping, 1– 12. https://doi.org/10.1002/hbm.25728
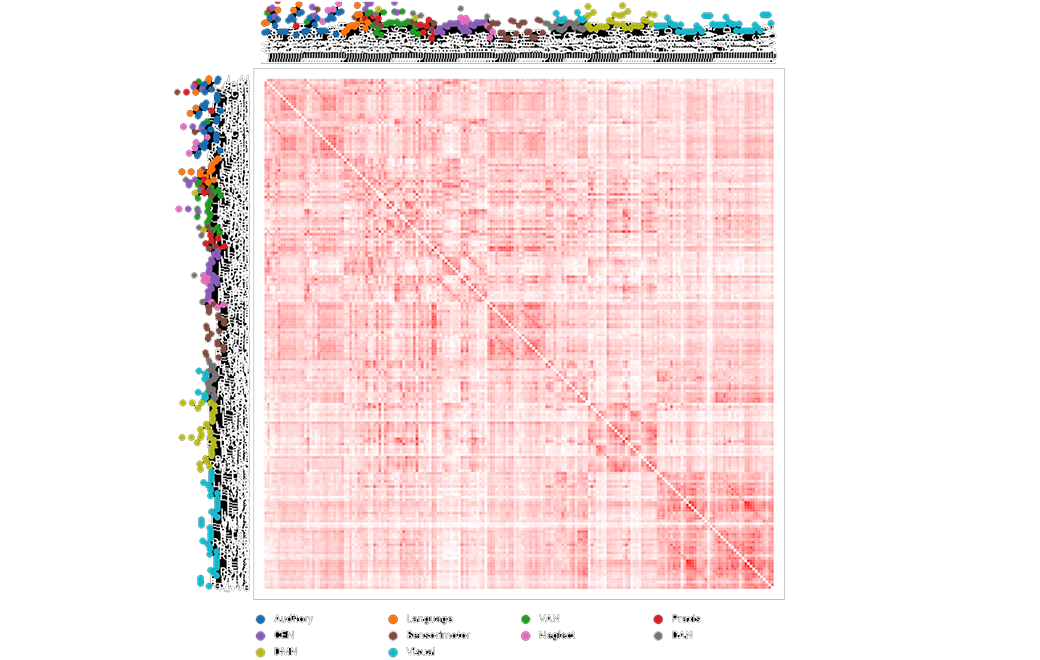
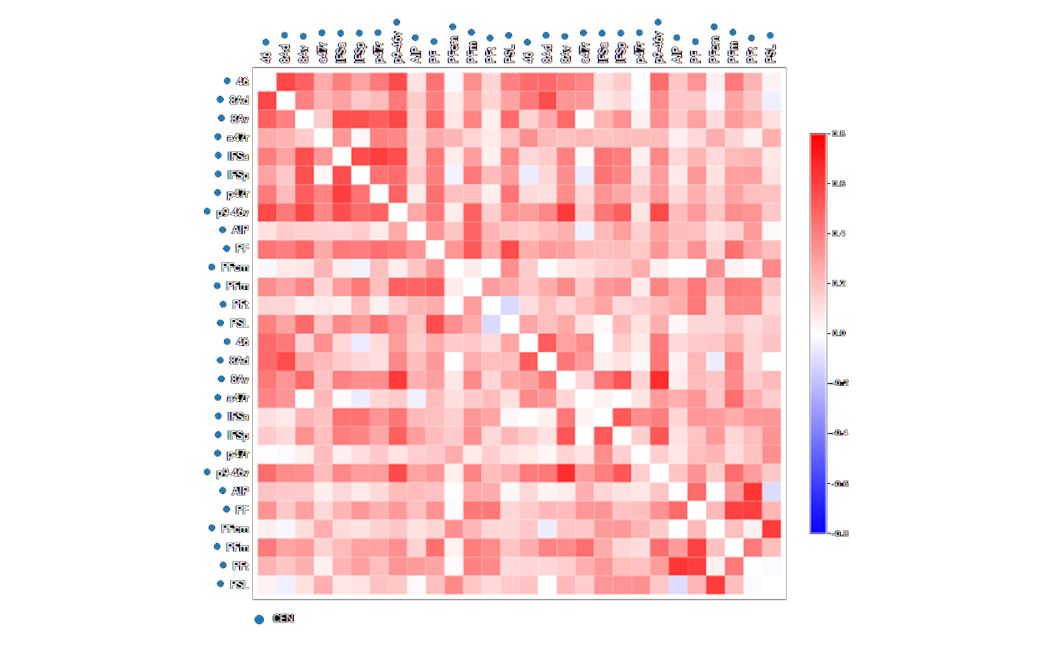
The connectivity between parcellations are measured, representing over 100,000 data points.
Through a simple selection, a brain network can be selected and viewed.
Pictured here, the subject’s Central Executive Network (CEN) in 3D.
The connectivity analysis is then focussed on relevant areas.
Red - positively correlated
Blue - negatively correlated
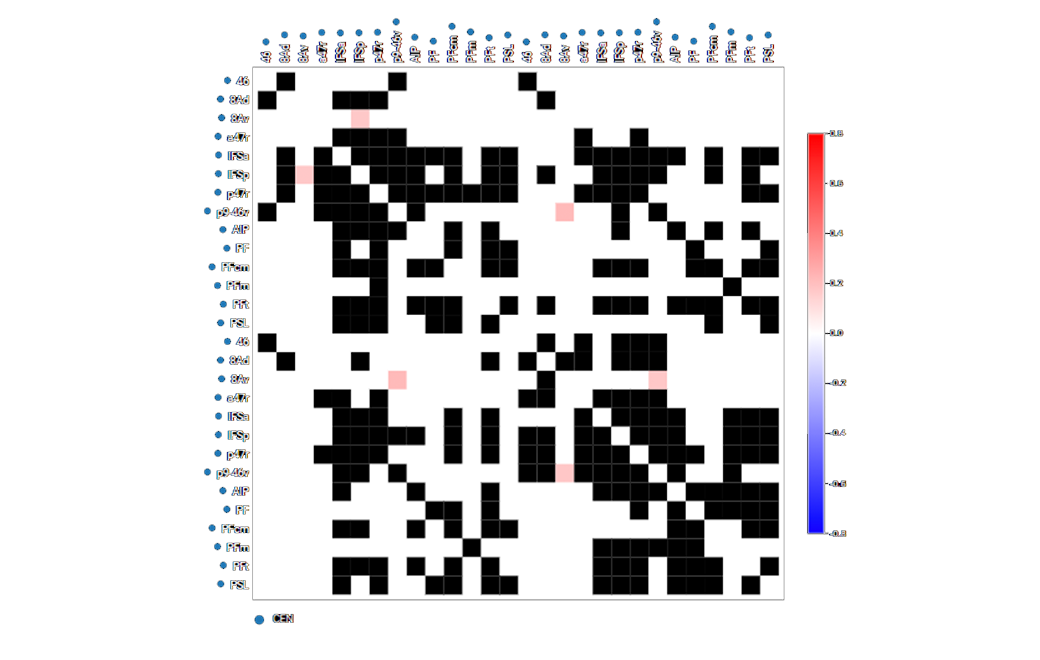
Next we use machine learning to determine areas of brain activity deviating from normal brain connectivity (as determined by user defined levels of standard deviations from the normal brain connectivity.)
Hypoconnectivity in blue
Hyperconnectivity in red
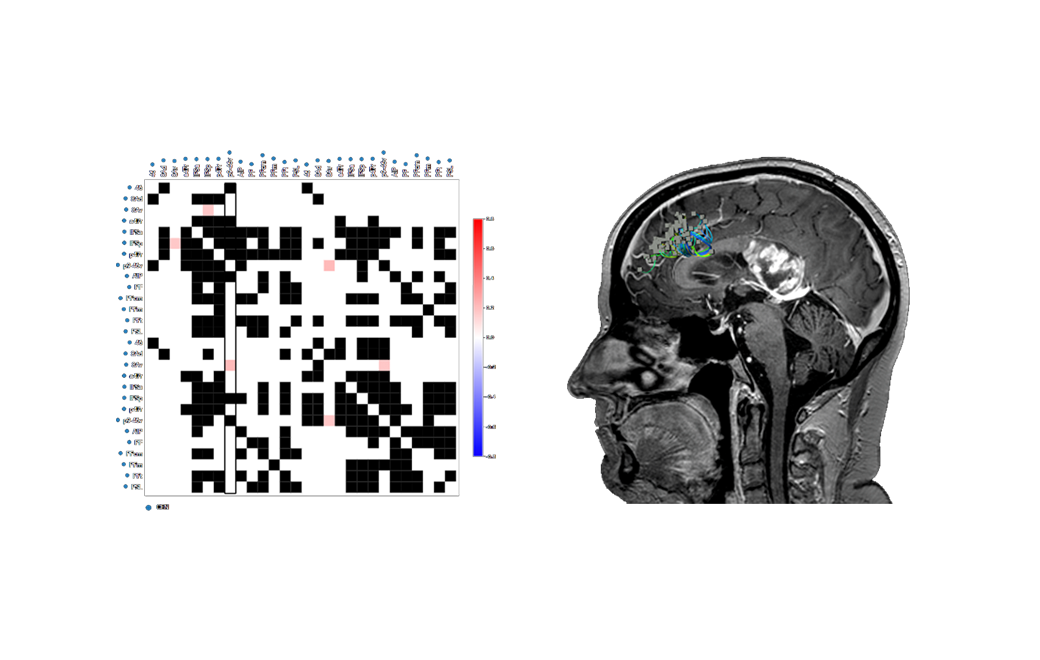
By selecting a region of interest, the software automatically co-registers this back onto the anatomical view.
In a few simple steps, we have now found a region of both functional and structural interest that may be exported and further investigated.
Discover our products for Neurologists & Researchers
Find out more today about how Omniscient is turning big data of the brain into actionable insights.


Products shown in this webpage have not been approved as medical devices or to support clinical decisions. Safety and effectiveness have not been reviewed by any regulatory agencies.

